| Table of Contents |  |
|
Original Article
| ||||||
| Effects of HIF-1α on ERRα/γ protein expression in mouse skeletal muscle | ||||||
| Ying Zhang1, Weixiu Ji1, Lianfeng Zhang2, Sixue Liu1, Gang Liu3, Jianxiong Wang4 | ||||||
|
1Institute of Sports Science, Beijing Sport University, China.
2Key Laboratory of Human Disease Comparative Medicine, Ministry of Health, Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences & Comparative Medical Center, Peking Union Medical College, China. 3Teaching Laboratory Centre, Beijing Sport University, China. 3School of Health and Wellbeing, Faculty of Health, Engineering, and Sciences, University of Southern Queensland, Australia. | ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
[Print This Article]
[Similar article in Pumed] [Similar article in Google Scholar] |
| How to cite this article |
| Zhang Y, Ji W, zhang L, Liu S, Liu G, Wang J. Effects of HIF-1α on ERRα/γ protein expression in mouse skeletal muscle. Edorium J Biomed Technol 2015;1:4–10. |
|
Abstract
|
|
Aims:
HIF-1α plays an important role in the adaptive responses to hypoxia. The ERRα and γ are crucial regulators of energy metabolism in skeletal muscle. The aim of the present study was to generate the inducible HIF-1α transgenic mice and examine the effects of different HIF-1α protein expression levels on ERRα/γ in mouse skeletal muscle.
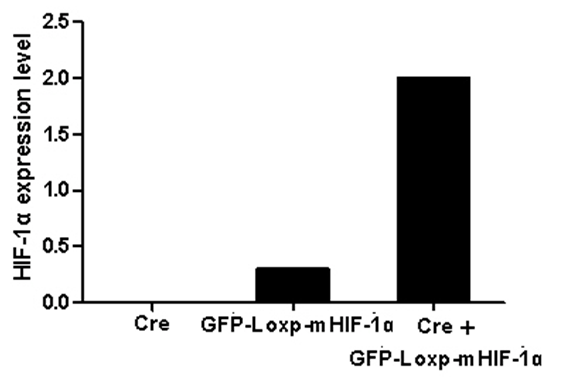
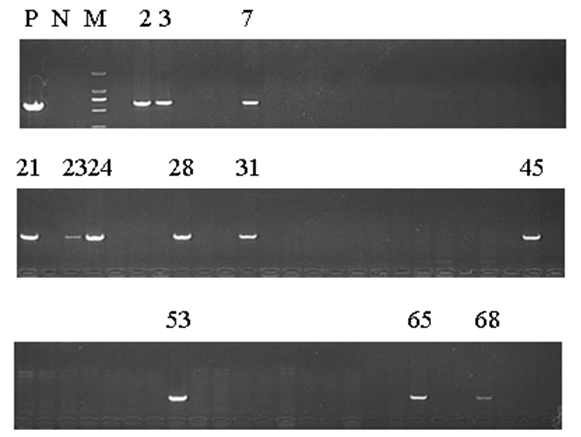
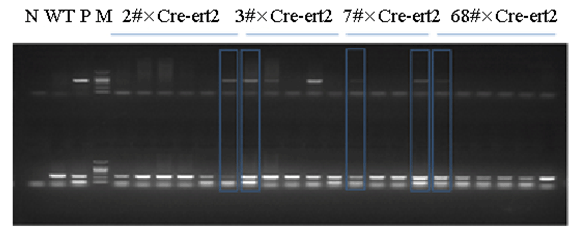
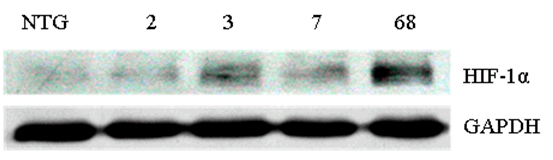
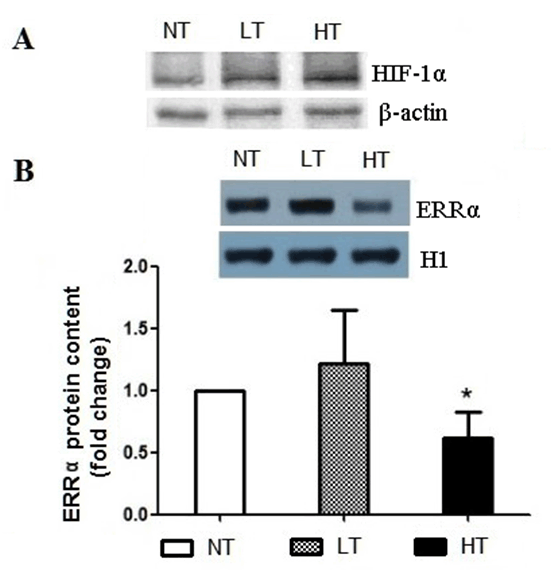
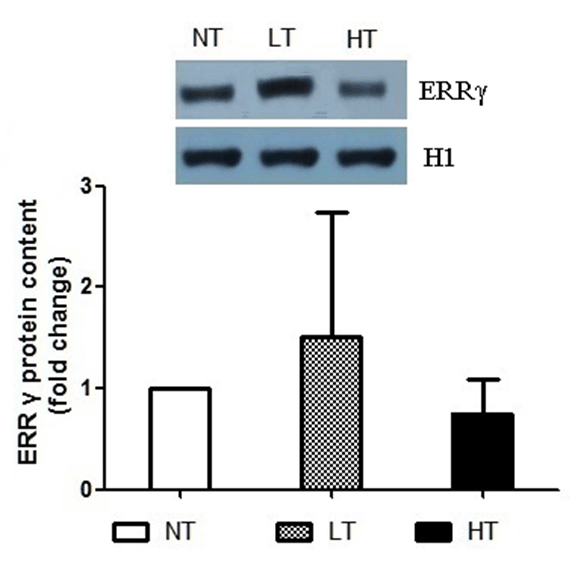
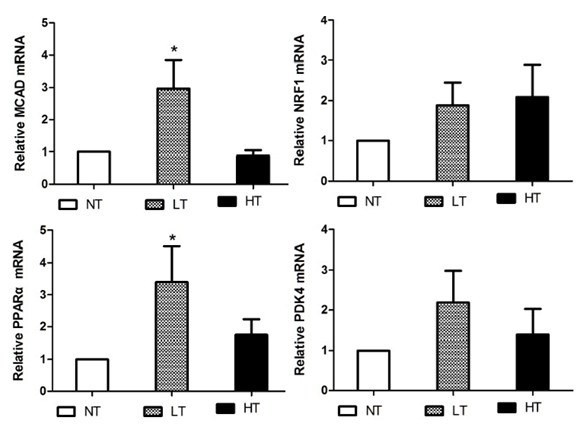
Methods: We generated the HIF-1α high-expression transgenic mice (HT) and HIF-1α low-expression transgenic mice (LT), and then compared the expressions of ERRα/γ and its target genes in skeletal muscles of three kinds of mice: HT, LT, and non-transgenic mice (NT). Results: The results showed that (1) the double positive mice from the founder of 3# and 68# showed an obvious expression of HIF-1α induced by tamoxifen and both of them were maintained to the further research as HT and LT mice, respectively; and (2) the nucleoprotein expressions of ERRα/γ and the mRNA levels of the ERRα/γ target genes: MCAD, PPARa, NRF1 and PDK4 were higher in the LT mice than the values in the NT, but only the mRNA levels of MCAD and PPARa were significantly higher. The HT mice showed significantly lower ERRα protein content than that of the NT mice. Conclusion: Our study was the first to report the generation of the inducible HIF-1α transgenic mice and effects of HIF-1α on ERRα/γ protein expression in mouse skeletal muscle in vivo. These data demonstrate that the low HIF-1α protein expression may associate with an up-regulation of ERRα/γ and their target genes in skeletal muscles, while the aggravated HIF-1α protein expression would reduce the effects. | |
|
Keywords:
ERRα/γ, HIF-1α, Skeletal muscle, Target genes, Transgenic mouse
| |
|
Introduction
| ||||||
|
The hypoxia inducible factor (HIF)-1 plays a key role to mediate the cellular responses to hypoxia. Its target genes control many cellular signal transductions, such as erythropoietin which induces red blood cell production, vascular endothelial growth factor (VEGF) which promotes angiogenesis, and glucose transporter 1 (GLUT1) which increases the efficiency of the glucose uptake [1] [2]. Previous studies have examined the role of HIF-1α in skeletal muscle. A research group found that the HIF-1 protein and mRNA levels are varied in different fiber types of rodent skeletal muscles [3]. Another study demonstrated that acute exercise can elevate the HIF-1 protein level in human muscle [4]. Deletion of HIF-1 in mouse skeletal muscle might lead to impaired glycolytic flux and decreased exercise endurance [5]. These studies focused on the effects of HIF-1 on the tissue or organ level. Skeletal muscle function and metabolism are controlled by many signaling pathways and molecules. At the molecular level, there are few studies about the relationship between HIF-1α and other signaling pathways or molecules which control energy metabolism in skeletal muscle. Estrogen-related receptors (ERRs) are crucial regulators of energy metabolism [6]. There are three members in the ERR family: ERRα, ERRβ and ERRγ. They are active nuclear receptors that contain high levels of the DNA sequence identified to that of estrogen receptors [7]. ERRα and γ isoforms are ubiquitously expressed at high levels in the heart, skeletal muscle and brown adipose tissue; while ERRβ expression is restricted to the brain, kidney and heart. ERRs control many downstream target genes which are important to glycolytic metabolism, fatty acid oxidation, mitochondrial oxidative phosphorylation, and mitochondrial biosynthesis. The main downstream target genes include pyruvate dehydrogenase kinase 4 (PDK4) [8] , medium-chain acyl coenzyme A dehydrogenase (MCAD) [9], peroxisome proliferator-activated receptor a (PPARa) [10], and nuclear respiratory factors (NRFs) [11]. There are a few reports about the relationship between HIF-1 and ERRs in cell lines in literature. A previous study has shown that ERRγ mRNA and protein levels were increased by a hypoxic treatment in the hepatoma cell lines and this change in ERR& #947; was associated with the increased HIF-1α and β expression [12]. The knockdown of endogenous HIF-1α reduced the hypoxia-mediated induction of ERRγ. In addition, hypoxia also increased the promoter activity and mRNA level of PDK4 in the HepG2 cells [12]. Another study suggested that ERRs physically interact with HIF and stimulate HIF-induced transcription. Transcriptional activation of hypoxic genes in cells cultured under hypoxia was largely blocked by suppression of ERRs through the treatment with diethylstilbestrol, an ERR inhibitor [13]. These results have suggested a possible relationship between HIF-1α and ERRs. However, these observations are restricted to the cell culture studies. There are few researches of the relationship between HIF1a and ERRα/γ in vivo. The purpose of this study was to generate the inducible HIF-1α transgenic mice and evaluate the effects of HIF-1α expression on the ERRα/γ protein and the mRNA expression levels of the ERRα/γ's target genes in skeletal muscle of the mouse with low- or high-expression in HIF-a protein. | ||||||
|
Materials and Methods
| ||||||
|
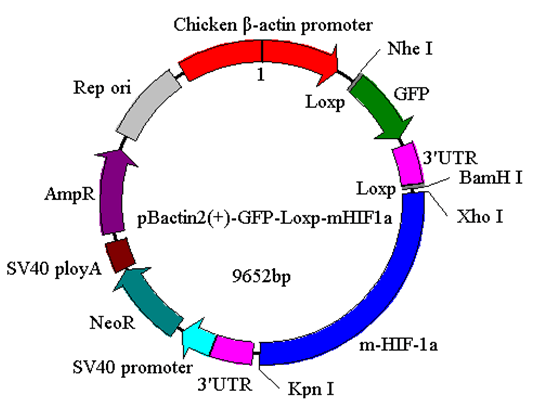
The generation of the inducible HIF-1α transgenic mice The present study protocol was approved by the Animal Care and Use Committee of Beijing Sport University. Non-transgenic mice (NT) (n=10), LT mice (n=10), and HT mice (n=10) were used to evaluate the effects of HIF-1α expression on the ERRα/γ protein and the mRNA expression levels of the ERRα/γ's target genes in skeletal muscle of the mouse. All mice with a mean body weight of 20±2 g were housed with controlled room temperature (20–25°C), 12:12-h light-dark cycle, and free access to food and water. After allowing acclimatization to their housing for three days, all mice were euthanized and their skeletal muscles from the legs were excised, cleaned of blood and connective tissue, quick-frozen with aluminum tongs, pre-cooled in liquid nitrogen, and stored at -80°C. Western blot analyses Real-time PCR Statistical analysis | ||||||
| ||||||
| ||||||
| ||||||
|
| ||||||
| ||||||
|
Results | ||||||
|
The generation of the inducible HIF-1α transgenic mice HIF-1α and nucleoprotein ERRα expressions in skeletal muscle The nucleoprotein expression of ERR& #947; in skeletal muscle The mRNA expression of MCAD, NRF1, PPARa and PDK4 in skeletal muscle | ||||||
| ||||||
| ||||||
| ||||||
|
| ||||||
| ||||||
|
Discussion
| ||||||
|
The importance of HIF-1 signaling in metabolic regulation has spurred much interest in recent years [12] [16]. Previous studies have been mainly focused on the metabolic role of HIF in
There are few studies referred to the relationship of HIF and ERR. Studying the effects of HIF-1α on ERRα/γ would improve our understanding about the metabolic regulation under hypoxic condition in skeletal muscle. Therefore, in the present study for the first time, we generated the inducible HIF-1α transgenic mice and reported the impact of HIF-1α on ERRα/γ protein expression in mouse skeletal-muscle through the new generated HIF-1α transgenic mice. There are no conclusive results of the effects of HIF-1 on ERRs in hypoxia. Some studies have reported that hypoxia stimulates the expression of ERRα through peroxisome proliferator-activated receptorγcoactivator-1α (PGC-1α), but not HIF-1. [21] [22]. While another study showed that HIF-1α had an effect on ERRs in hypoxic cell culture in vitro [12]. This statement has been supported by the finding of two putative binding sites of HIF-1α on the ERR& #947; promoter in a cell culture experiment. The promoter activity in response to hypoxia was abolished in these site-mutated constructs [12]. Furthermore, the evidence of ERR& #947; regulation of HIF-1α under hypoxic condition was confirmed by the ChIP assays in the hypoxia treated HepG2 [12]. In the present study, we observed that the nucleoprotein expressions of ERRα/γ and the mRNA levels of the ERRα/γ target genes: MCAD, PPARa, NRF1 and PDK4 were higher in the LT mice compared to the values in the NT mice, although only the mRNA levels of MCAD and PPARa were increased significantly. The results suggest that HIF and ERR may act in a common pathway and form a regulatory complex. Interestingly, we also found that the ERRα/γ protein contents were decreased in the HT mice and the reduced ERRα was significantly different from the corresponding NT value. HIF-1α is able to induce apoptosis during severe or prolonged (>24 hours) hypoxia, although it has been generally accepted as a pro-survival factor [13] [23]. Thus, the molecular mechanism causing different ERRα/γ protein levels, transcriptional regulation or output of ERRα/γ for downstream targets requires further clarification. There are limitations in our study, we had no single transgenic mice of GFP-Loxp-mHIF-1a and Ubc-Cre-ert2 as controls. More controls would provide more clear results to the research. In addition, we did not measure the transcriptional activity of the ERRα/γ genes which are regulated by HIF-1α, as well as the ERRα/γ mRNA level and the transcriptional activity of PDK4, MCAD, PPARa and NRF1 genes which are regulated by ERRα/γ. Though the present results have shown the effects of HIF-1α on the expressions of ERRα/γ and their target gene, future studies should measure these directly. Moreover, in the present study, we only investigated the gene and protein expressions but did not do any histomorphological and functional measurements. These histomorphological and functional measurements would be desirable and add more comprehensive insights into the scope of the study. | ||||||
|
Conclusion
| ||||||
|
Our study was the first to report the generation of the inducible HIF-1α transgenic mice and effects of HIF-1α on ERRα/γ protein expression in mouse skeletal muscle in vivo. We found that the nucleoprotein expressions of ERRα/γ and the mRNA levels of the ERRα/γ target genes were higher in the LT mice; while the ERRα/γ protein contents were decreased in HT mice, compared to the values in the NT mice respectively. These data reveal a relationship that HIF-1α affects ERRα/γ and their target genes in mouse skeletal muscle. | ||||||
|
Acknowledgements
| ||||||
|
This study was funded by grants from the National Natural Science Foundation of China (31471134; 31171140) and Program of General Administration of Sport of China (2014B010). | ||||||
|
References
| ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
|
|
Author Contributions:
Ying Zhang – Substantial contributions to conception and design, Acquisition of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Weixiu Ji – Substantial contributions to conception and design, Acquisition of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Lianfeng Zhang – Substantial contributions to conception and design, Acquisition of data, Drafting the article, Final approval of the version to be published Sixue Liu – Substantial contributions to conception and design, Acquisition of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Gang Liu – Substantial contributions to conception and design, Acquisition of data, Drafting the article, Final approval of the version to be published Jianxiong Wang – Substantial contributions to conception and design, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published |
|
Guarantor of submission
The corresponding author is the guarantor of submission. |
|
Source of support
None |
|
Conflict of interest
Authors declare no conflict of interest. |
|
Copyright
© 2015 Ying Zhang et al. This article is distributed under the terms of Creative Commons Attribution License which permits unrestricted use, distribution and reproduction in any medium provided the original author(s) and original publisher are properly credited. Please see the copyright policy on the journal website for more information. |
|
|