| Table of Contents |  |
|
Editorial
| ||||||
| The conservation of genotoxic stress-induced morphological changes in yeasts | ||||||
| Jia-Ching Shieh1,2 | ||||||
|
1Associate Professor, Laboratory of Unicellular Eukaryotes, Department of Biomedical Sciences, Chung Shan Medical University, Taichung City, Taiwan, Republic of China.
2Department of Medical Research, Chung Shan Medical University Hospital, Taichung City, Taiwan, Republic of China. | ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
[Print This Article]
[Similar article in Pumed] [Similar article in Google Scholar] |
| How to cite this article |
| Shieh Jia-Ching. The conservation of genotoxic stress-induced morphological changes in yeasts. Edorium J Biomed Sci 2015;1:1–6. |
|
The Conserved Pathways for Genome Integrity Induced by Genotoxic Stress
| ||||||
|
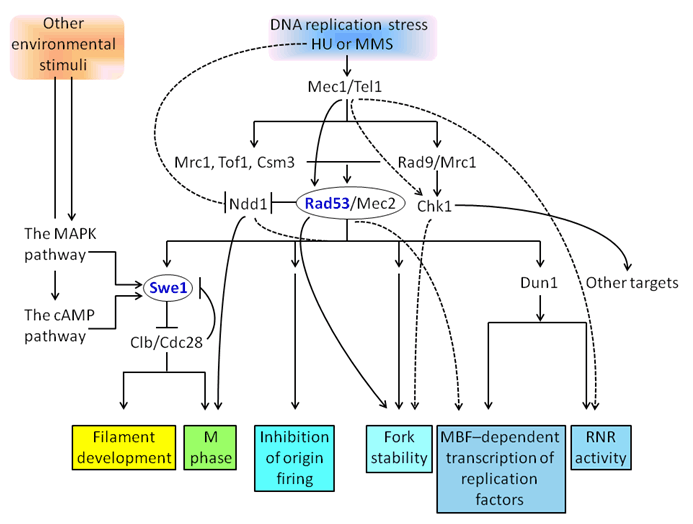
Genomic stability is vital to sustain cell survival and to prevent errors in control of proliferation that often result in disease. To maintain genomic stability, highly conserved signaling pathways of DNA integrity checkpoints are required to ensure the activation of the pathways to delay cell cycle progression until DNA repair or replication is complete after genotoxic stress that leads to DNA damage or replication blocks. Much of the work that has deciphered the DNA integrity checkpoints is from the study of the budding yeast Saccharomyces cerevisiae. At least two checkpoints are known to operate during S phase in budding yeast: a replication checkpoint, originally observed after a hydroxyurea (HU)-induced deoxynucleoside triphosphate depletion, which blocks the progression of S phase in the replicative forks from both early- and late-firing origins [1], and an intra-S phase checkpoint, which lowers the rate of DNA replication and slows cell cycle advance in response to DNA-damaging agents [2] [3]. Additionally, checkpoints that operate to censor the assembly of the mitotic spindle and to control progression through mitosis have been discovered [4]. Central to the DNA integrity checkpoints are the highly conserved S. cerevisiae PI3K -related protein kinases Tel1 and Mec1 (Figure 1), the mammalian homologs of ATM and ATR [5] [6]. When DNA damage is detected by sensor proteins Mec1 and Tel1, which signal through mediator Rad9 to downstream effectors Rad53 and Chk1 [7] to cause mitotic arrest in the G1 and G2 phases [8] [9][10], three components of the DNA replication machinery-Mrc1, Tof1, and Csm3-operate as the replication checkpoint mediators as opposed to Rad9 [8] [11][12] (Figure 1). Thus, during normal DNA replication, these mediators function differently from when they are activated as part of a checkpoint [11] [12][13][14][15][16][17]. Most recently, Ndd1, a new component whose activity is controlled by genotoxic stress, has been identified [18] [19][20]. During methyl methane sulfonate (MMS)-induced G2/M arrest, Ndd1 activity is blocked exclusively in a Mec1-Rad53-dependent manner. HU induces a cell cycle arrest that prevents cell cycle progression into S phase and inhibits Ndd1 activity by Mec1-Rad53 through unknown mechanisms (Figure 1). Unlike the fission yeast Schizosaccharomyces pombe and higher eukaryotes whose cell cycle progression is paused in response to replication stress, principally by promoting inhibitory phosphorylation of the cyclin-dependent kinase (CDKs), S. cerevisiae blocks cell cycle progression by directly inhibiting origin firing and chromosome segregation [1] [21] | ||||||
|
Genotoxic stress-induced morphological alterations in the model yeast S. cerevisiae
| ||||||
|
In S. cerevisiae, Cdc28, the mammalian equivalent of Cdk1, along with its associated cyclins (Clbs), appears to coordinate the cell cycle with bud morphogenesis [22][23] [24]. It seems reasonable that the checkpoints of morphogenesis and those of DNA integrity are interconnected to allow for the tight coordination of cell cycle advancement and morphogenesis because the presence of morphogenesis checkpoints permits the cell to monitor defects in bud morphology, bud formation, septin organization, cell size, and cell wall synthesis, as well as perturbations in the actin cytoskeleton [23] [25] [26][27][28]. Swe1, a S. cerevisiae homolog of mammalian and S. pombe Wee1, is central to the morphogenesis checkpoints [23]. In budding yeast, Swe1 phosphorylates and inhibits Clb-bound Cdc28 on the Tyr19 residue [29] (Figure 1), a modification that is abolished by Mih1, the mammalian and S. pombe Cdc25 homolog [30][31]. During a normal cell cycle, Swe1 accumulates in the S phase, becomes serially hyperphosphorylated [32] [33], producing multiple isoforms, and undergoes ubiquitin-mediated degradation [31] [34][35][36]. Defects in septin filament assembly at the bud neck [32] [37][38][39][40], as well as perturbations in the cell size, bud formation, and the actin cytoskeleton [23] [25], cause hypophosphorylation and stabilization of Swe1 and, consequently, the Swe1-dependent inhibition of Clb-Cdc28. This Swe1-imposed G2 delay leads to elongated cells, as Clb-Cdc28 cannot induce the switch from polarized to isotropic growth during budding [23] [41][42] (Figure 1). Swe1, a S. cerevisiae homolog of mammalian and S. pombe Wee1, is central to the morphogenesis checkpoints [23]. In budding yeast, Swe1 phosphorylates and inhibits Clb-bound Cdc28 on the Tyr19 residue [29] (Figure 1), a modification that is abolished by Mih1, the mammalian and S. pombe Cdc25 homolog [30][31]. During a normal cell cycle, Swe1 accumulates in the S phase, becomes serially hyperphosphorylated [32, 33], producing multiple isoforms, and undergoes ubiquitin-mediated degradation [31] [34][35][36]. Defects in septin filament assembly at the bud neck [32] [37][38][39][40], as well as perturbations in the cell size, bud formation, and the actin cytoskeleton [23] [25], cause hypophosphorylation and stabilization of Swe1 and, consequently, the Swe1-dependent inhibition of Clb-Cdc28. This Swe1-imposed G2 delay leads to elongated cells, as Clb-Cdc28 cannot induce the switch from polarized to isotropic growth during budding [23] [41][42] (Figure 1). Several other Swe1 regulators have also been shown to influence morphogenesis. Hsl1, the S. pombe Nim1/Cdr1, is a primary negative regulator of Swe1 and is required for efficient Swe1 localization at the bud neck [24][37][43]. UnlikeNim1/Cdr1, which is capable of phosphorylating Wee1, Hsl1 is unable to phosphorylate Swe1 [44]. Cla4/PAK and Cdc5/Polo, which are sequentially targeted to the neck of bud, appear to be responsible for the stepwise phosphorylation and down-regulation of Swe1 [45]. Additionally, Hsl1 appears to function in concert with Hsl7 [32] [46][47], as the absence of either Hsl1 or Hsl7 radically lowers Swe1 phosphorylation in vivo and results in cell elongation. However, the finding that the need for the Hsl1-Hsl7 interaction in Swe1 degradation can be bypassed by tethering Swe1 to the septins provides evidence that Hsl1-Hsl7 has a downstream role involving the presentation of Swe1 to other regulators for targeting to the degradation pathway [48]. Moreover, Swe1 degradation is regulated by its interaction with Clb2-Cdc28, Cdc5/Polo, and Hsl1. Swe1, synthesized during the S and G2 phases, binds to Clb2-Cdc28, where it is protected from Cdc5-specific phosphorylation. In late G2, when the levels of Hsl1 and Cdc5 rise, Hsl1 leads to the dissociation of Swe1 from Clb2-Cdc28, enabling Cdc5 to phosphorylate Swe1, which leads to its ubiquitin-mediated degradation [49] [50]. Recently, it has been found that feedback between Swe1 and Cdc28 controls the Swe1 abundance following stress. Swe1 inhibits Cdc28, which in turn antagonizes Swe1 by promoting its transcriptional repression and its degradation. In cells with mature septin rings, stresses due to osmotic shock or actin depolymerization promote the ability of Swe1 to inhibit Cdc28 but do not directly stabilize Swe1, resulting in subsequent stabilization and accumulation of Swe1 via feedback [38] (Figure 1). A positive feedback loop in which Swe1 activity inhibits the CDK, which then ceases to target Swe1for degradation, is of physiological importance. In S. cerevisiae, various conditions that slow DNA synthesis are responsible for the induction of filamentous differentiation through Mec1-Rad53-Swe1-Cdc28-Clb2, as has been demonstrated earlier [51]. Under restrictive temperatures, DNA polymerase and DNA ligase temperature-sensitive mutants induce filamentous growth. In addition to HU, MSS, a DNA-alkylating agent that can slow DNA replication fork progression [52], and ara-CMP, a potent inhibitor of yeast DNA polymerases [53], induce filamentous growth. Since the MAPK and cAMP signaling pathways mediate nitrogen starvation-induced filamentous growth in S. cerevisiae, their possible involvement in genotoxic stress-induced filamentous growth has been investigated. As FLO8 encodes a DNA-binding transcriptional regulator that serves as the effector for the cAMP pathway and TEC1 encodes a DNA binding transcriptional regulator that serves as the effector for the MAPK pathway, tec1?flo8D mutant cells defective in both signaling pathways were created to examine their responses to genotoxic stress. Cells of the tec1?flo8D mutant respond to genotoxic stress with normal filamentous growth [51]. Additionally, HU-induced filamentous growth shows dependence on Swe1 that is essential for nitrogen starvation-induced filamentous growth [51]. Moreover, DNA-damaging agents such as bleomycin, etoposide, and H2O2 fail to induce filamentous growth despite the fact that the Mec1-Rad53 DNA integrity checkpoint proteins can also be activated by DNA damage via the Rad9-Chk1-dpendent DNA damage response checkpoint [51], demonstrating that the Rad9-Chk1-dependent DNA checkpoint pathway is not required for the Mec1-Rad53-mediated filamentous growth. Taken together, DNA replication stress-induced filamentous growth, mediated through Mec1-Rad53-Swe1-Cdc28-Clb2, and nitrogen starvation-induced filamentous growth, mediated by the MAPK and cAMP signaling pathways, converge at Swe1 (Figure 1). | ||||||
|
| ||||||
| ||||||
|
The divergent pathways for genotoxic stress-induced morphogenesis in the fungus C. albicans
| ||||||
|
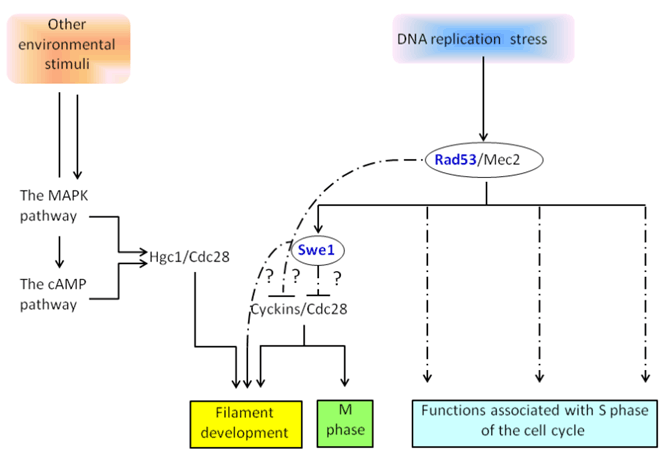
It is of great interest to know if the genotoxic stress-induced filamentous growth is conserved in other fungi, particularly the important opportunistic human fungal pathogen Candida albicans [54] [55], whose ability to switch between the yeast and filamentous forms (pseudohyphae or hyphae) is critical to its virulence [56]. Upon response to environmental cues such as temperature, serum, and pH, C. albicans can change from a yeast (isotropic) form of growth to a filamentous (polarized) type growth. Under genotoxic stresses, such as HU and MMS, C. albicans can also be triggered to initiate polarized growth [57] [58], suggesting a conservation between C. albicans and the budding yeast in genotoxic stress-induced filamentous growth. Importantly, hydrogen peroxide activates hyphal development through Rad53 [59], whereas deleting RAD53 abolishes genotoxic stress-induced filamentous growth in C. albicans [60]. In contrast, inactivation of mitotic recombination proteins such as Rad50, Rad51, Rad52, and Mre11 or Cln3, Clb2, or Clb4 cyclins stimulates constitutive polarized growth in C. albicans [57] [60] [61][62][63][64]. This implies that the DNA integrity network and cell cycle proteins may represent new regulators of filamentous growth in C. albicans . Interestingly, through the site-directed mutagenesis of Rad53, it was found that the functions of Rad53 in DNA repair and replication arrest can be separated from its role in genotoxic stress-induced polarized growth in C. albicans [58] [60][65] It has been known that the Hsl1-Swe1-Cdc28 pathway is important for cell elongation of both the yeast and hyphal forms and for virulence in C. albicans [66]. However, the phosphorylation state of Tyr19 on Cdc28 between yeast and hyphal cells does not appear to be different [67], indicating that Tyr19 phosphorylation on Cdc28 may not be important for polarized growth in C. albicans and that Swe1, which phosphorylates Tyr19 on Cdc28, is not required for hyphal growth. Essentially, even though yeast cells lacking SWE1 are slightly rounder in shape than wild type cells, they form normal pseudohyphae and hyphae [68]. Cell cycle delays in response to DNA damage leading to polarized growth are partially dependent upon Swe1 [61] . These results suggest that genotoxic stress-induced filamentous growth is not or is only partially mediated by Swe1, unlike the Rad53-Swe1-dependent process observed in S. cerevisiae. It will be interesting to see if cells lacking SWE1 abolish genotoxic stress-induced filamentous growth in C. albicans. Intriguingly, DNA damage-induced filamentous growth involves but does not require the expression of hyphal-specific genes or the Cph1 and Efg1 transcription factors, which are downstream targets of the MAPK and cAMP signaling pathways, respectively [61]. Additionally, cells lacking both CPH1 and EFG1 can be induced to filamentous growth in response to HU [69]. Particularly, filastatin, a small molecule, has been found to block hyphal growth induced by serum, Spider media, and GlcNac but not by the genotoxic agent HU [70]. Filastatin inhibits the transcriptional activation of HWP1 [70], which is an early and essential event in the process of hyphal development [71]. Taken together, there may be a pathway that is dependent on Rad53 but independent of Swe1 and the MAPK/cAMP signaling pathways for the induction of filamentous growth in C. albicans (Figure 2). The rewiring of the genotoxic stress-induced filamentous growth pathway may be associated with the interaction of C. albicans with its host. Determination of the differences between the S. cerevisiae and C. albicans on the control of genotoxic stress-induced filamentous growth is important and has a potential in therapeutics. Keywords: Yeast, DNA integrity checkpoint, Genotoxic stress, DNA replication stress, Morphogenesis | ||||||
|
| ||||||
| ||||||
|
Acknowledgements
| ||||||
|
This work was supported by the grants from the Ministry of Science and Technology of Taiwan, Republic of China (NSC 97-2320-B-040-014-MY3 and NSC 101-2629-B-040-001-MY3) and from the National Health Research Institutes of Taiwan, Republic of China (NHRI-EX99-9808SI & NHRI-EX100-9808SI) | ||||||
|
References
| ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
|
|
Author Contributions:
Jia-Ching Shieh – Substantial contributions to conception and design, Acquisition of data, Analysis and interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published |
|
Guarantor of submission
The corresponding author is the guarantor of submission. |
|
Source of support
None |
|
Conflict of interest
Authors declare no conflict of interest. |
|
Copyright
© 2015 Jia-Ching Shieh. This article is distributed under the terms of Creative Commons Attribution License which permits unrestricted use, distribution and reproduction in any medium provided the original author(s) and original publisher are properly credited. Please see the copyright policy on the journal website for more information. |
|
|